Neuron Simulation, notes
Table of Contents
Week 1 - Simulation Neuroscience: An Introduction
Understanding the brain
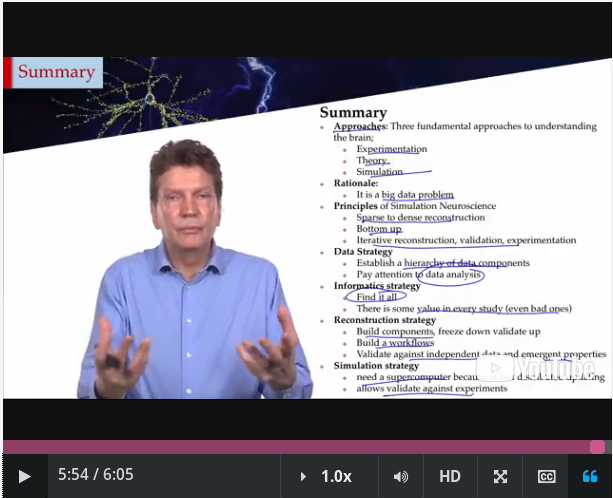
Approaches, which, in turns, following out three branches of exploratory science:
- Experimental,
- Theoretical
- Informatics neuroscience (looking at trend, correlations and patterns in data),
- Theoretical neuroscience (explaining the data and predicting
- Computational neuroscience (creating minimal model that fit specific experimental data),
- Applied neuroscience (using neurosciences in medicine and engineering by notably building machine to see how the neurons work)
- & Simulation (or
lqqdthat this course is about).
Principles:
- Dense reconstructions (it's about an ordered system so, opposite than it were about a random system) it's assumed to get done algorithmically (the smallest data set with & the [algorithm|s] which [rule|s] over).
- Bottom-up reconstructions,
- Iteratively reconstructions & test.
Experimental data
I am going to track this exploration on the subject on a handful of idea tools as keystrokes, connections | flows & systems | circuits.
- So, my first annotation here is the flow in which this approach about neuron as a whole is on, ion channels, synapses & neuron circuits.
- Now lets track the info source in terms of simulation so we can control also the replication method (rationale & the complexion itself got made)
ion channelchannel screening, biophysical screening so the explore the biophysics of these channels.. implying kinetics (open closed) of the channels, its distribution, combination, conductance, so modelling the ion channel, its morphology and | or its electrical behaviour,neuronspaired recordings, staining the two..n. circuitsantibody staining, in situ hybridization,
The question quiz here is indeed my point when defining the keystroke specially about (simulating) neuron.
— What is, in your opinion, the main challenge in mining of existing knowledge on the brain. I feel the keystroke about mining is the coding language we opted to interpret the data and build to simulations. As a programming language is necessary, the question easily gets deviation to the interface or media used itself, its virtues, shortcomings and so on. Especially if researchers are previously versed in any of them. I do not think this fact were intrinsic wrong, researchers should be able to explain their approaches within plenty of comfort zones. However this fact triggers at least two inconveniences: First, it narrows the replication, or, the result evaluation or, lets also call it, the discussion phase, into groups based on the language media used to expose the results or its eventual approaches. How many times me myself, as student or, just as, an intelligent plain man interested in the field have had to just observe what exposed and without ability to replying because of the programming language barrier. So that as soon as you decide to get levelled-enough literacy to, in turn, get into that specific discussion some more interactive, reply based on whatever different language tool there were out, shows up. So that, the easy route this question ends up is a kind of school of exposition problem.
A second inconvenience is the virtues of the media or language in question itself. A lot of discussions as I found got suck as function of packages used to expose approaches.
I think we have to agree in a kind of standard, a kind of pseudo code as a default language so that any researcher were able to replicate results.
- Hey, —mmm… —when, a kind of, positive difference, should not be considered an advantage, is that like, as I should get it rendered this, kind of, dissonance? Look at this question [ref: week1, practice quiz :reconstruction strategies] or is it just another issue for my
WTFseries collection?
Caveats?
Let's list some of them.
- critical data may be missing
- biological data has a lot of mistakes (actually there are much more mistakes than accurate data)
- there are a lot of disciplines involved (which is not a caveat, really, though it get things complicated, critically complicated, actually).
Single neuron data.
Basically in search of: IR-DC microscope,
- its electrical profile
- morphological profile,
- molecular profile.
Extra Annotation.
Synapses
Graded Quiz 1: An introduction to Simulation Neuroscience
Graded week quiz This content is graded
Week 2 - Neuroinformatics
Learning goals Setup Introduction to neuroinformatics Knowledge graphs and ontologies Neuroinformatics data Acquisition of neuron electrophysiology and morphology Morphological feature extraction Graded Quiz 2: Neuroinformatics Graded week quiz This content is graded Graded assignment 1 Weekly graded assignment This content is graded
Week 3 - Modeling Neurons
Learning goals Introduction to the single neuron The neuron Electrical neuron model Tutorials creating single cell electrical models Graded Assignment 2 Weekly graded assignment This content is graded Graded quiz 3 Graded week quiz This content is graded
Week 4 - Modeling Synapses
Introduction and modeling synaptic potential Modeling synaptic transmissions between neurons Set up your Collab week 4 Tutorial: Modeling dynamic synapses Graded Assignment 3 Weekly graded assignment This content is graded Graded quiz 4 Graded week quiz This content is graded
Week 5 - Constraining Neuron Models with Experimental Data
Constraining neuron models with experimental data Set up you Collab week 5 Tutorials for optimisation Graded Assignment 4 Weekly graded assignment This content is graded Graded quiz 5 Graded week quiz This content is graded